Microsporidia
Microsporidia are known to infect half of all animal phyla as well as some protists. -Murareanu*, Sukhdeo*, Qu*,Jiang*, et al, .mBio, 2021.
Microsporidia are the earliest diverging fungi and obligate intracellular pathogens that can specifically infect a diverse range of different hosts, including a number of species that cause death and disease in humans and agriculturally important animal species.
Despite the ubiquitous nature of these pathogens, very little is known about them. To understand how microsporidia are able to successfully infect and proliferate inside of their hosts, the Reinke lab is taking advantage of several naturally occurring microsporidia species that infect experimentally tractable nematodes. This system has three critical advantages for studying pathogen evolution. 1) It is a natural system of evolutionary related coevolved pathogens that infect hosts that are genetically and biochemically tractable. 2) All infection stages can be studied entirely in vivo allowing physiologically relevant evolutionary studies not possible in other animal systems. 3) The related parasites exhibit different phenotypes including different host ranges and tissue tropisms.
Microsporidia Genomics
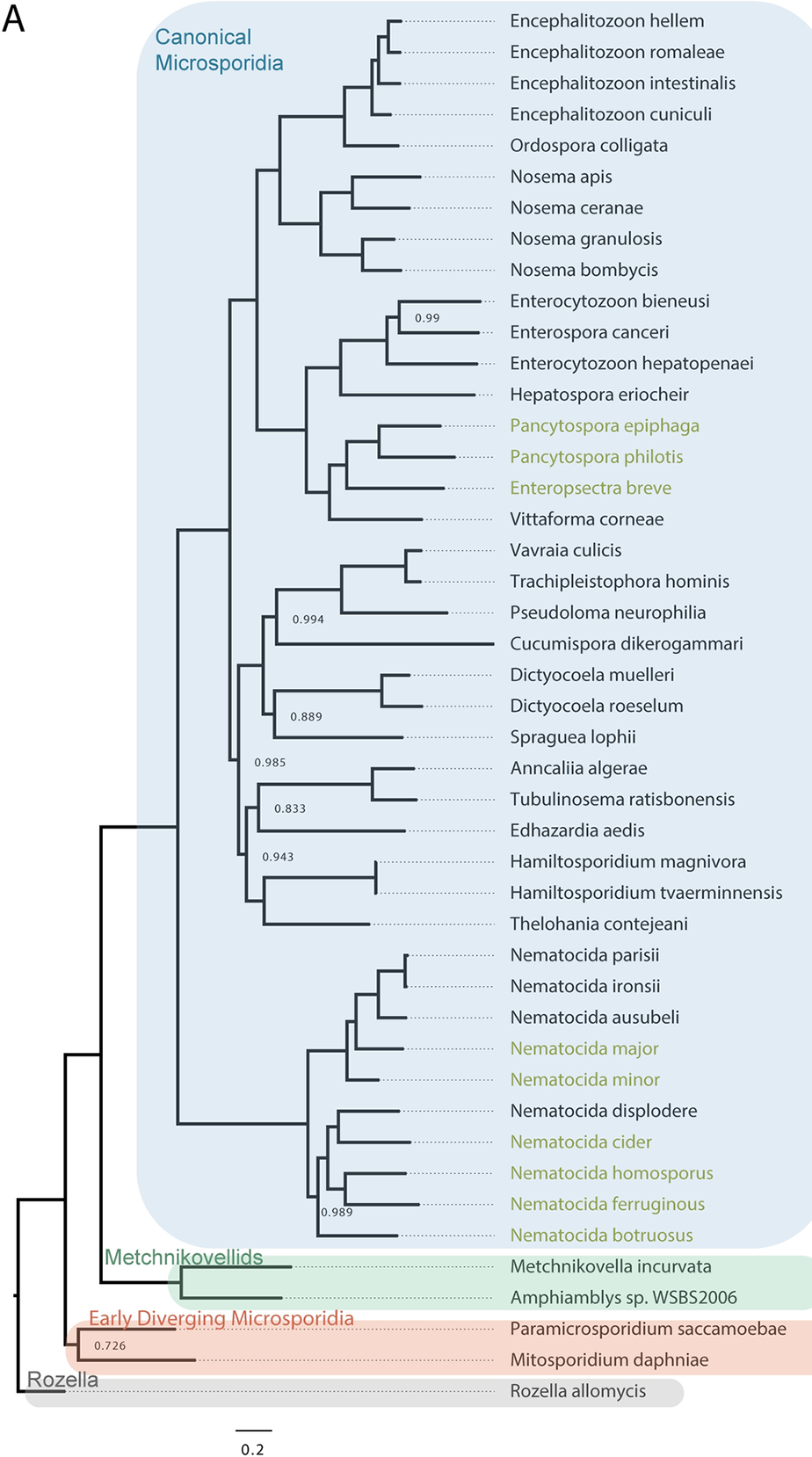
Phylogenetic tree of microsporidia and their close ancestors. - Wadi et al., Plos Pathogens, 2023
Microsporidia have the smallest known genomes of any eukaryotes with some species having only ~2000 protein coding genes. Microsporidia have rapidly evolved, containing only ~800 proteins that are conserved outside of microsporidia. These parasites are the “simplest” eukaryotes and, remarkably, as a phylum have evolved the capabilities to infect most types of animals. Over 1400 species of microsporidia have been identified and many species infect humans and agriculturally important animals including honeybees, silkworms, shrimp, and fish.
We previously employed a location-based proteomics technique to identify potential pathogen effector proteins that are localized within host intestinal cells, providing the first large-scale experimental localization of pathogen proteins inside cells of a living animal host.
We have performed genomic sequencing, analysis, and assembly of genomes from nematode-infecting microsporidia. There are similarities between these nematode infecting microsporidia, such as many metabolic pathways that are conserved. There are also differences between species, such as large, expanded families which are uniquely present in many species. Strikingly, one previously identified Nematocida family is present in Pancytospora species.
Using computational and experimental approaches, we have found that microsporidia proteins have undergone extensive reduction in both size and function.
Mechanisms of RESISTANCE against microsporidia
C. elegans parents that are infected with microsporidia produce progeny that are resistant to subsequent infection. - Willis AR*, Zhao W*, et al., Science Advances, 2021
C. elegans that are missing AAIM-1 are resistant to infection by N. parisii, but susceptible to the pathogenic bacteria P. aeruginosa. - Tamim El Jarkass H, et al., eLife. 2022.
Understanding the genetic basis of pathogen resistance is important for determining both the factors that the host uses to defend against the pathogen and the host factors that the pathogen is exploiting for its own benefit.
We are approaching this problem by performing screens to identify mutations that alter host resistance. We are also working on identifying naturally occurring mutations that are responsible for pathogen resistance. In addition to genetic mechanisms of resistance, we are also studying an intergenerational form of immunity. Parents infected with microsporidia produce resistant progeny, and this immunity is induced via a somatic transcriptional response in the parents. Parential nutrition also influences progeny as parents grown on bacteria containing vitamin B12 produce offspring that are tolerant of microsporidia infection.
Together this work will reveal how hosts evolve resistance against multiple related pathogen species and the extent to which pathogens influence host genome evolution.
microsporidia Inhibitors
Prevention of N. parisiii proliferation by the drug dexrazoxane.-Murareanu et al, Nature Communications, 2022
Microsporidia cause disease in humans as well as other animals such as honeybees, fish, and shrimp. In recent years, the number of reported infections has been increasing, but there are very limited therapeutic options. Because of this there is a pressing need to identify drugs that can be used to treat infections by these parasites. We have identified novel inhibitors of this pathogen by developing a novel screen platform using C. elegans infected with N. parisii. Infection with the microsporidian parasite prevents these worms from producing offspring, which provides a fast and convenient readout for discovering inhibitors. We screened ~2600 compounds for activity and identified 11 compounds that reduce the amount of pathogen in the animals. One of these identified compounds prevents the pathogen from growing, and several others prevents the pathogen from invading the host by inhibiting spore firing. Using our C. elegans-N. parisii assay, we have performed additonal screens to identify other inhibitors of microsporidia infection. Future work will further characterize these compounds, including identifying the microsporidia proteins that are being inhibited.